How to work with Taxon Framework Relationships
If a Taxon Framework has a source and covers downstream taxa, then it can have Taxon Framework Relationships. These taxon framework relationships are created and edited by iNat curators and describe the mapping between internal taxa on iNaturalist and the external taxa in the reference.
How to set them up
Imagine we have an Taxon Framework sourced to an external reference with three external taxa. Meanwhile, there are 5 internal taxa on iNaturalist covered by the Taxon Framework.
The next step is to account for the external taxa by wiring them up to the external taxa with Taxon Framework Relationships. This can be automated to some extent, but to do it correctly, things need to be manually inspected an curated.
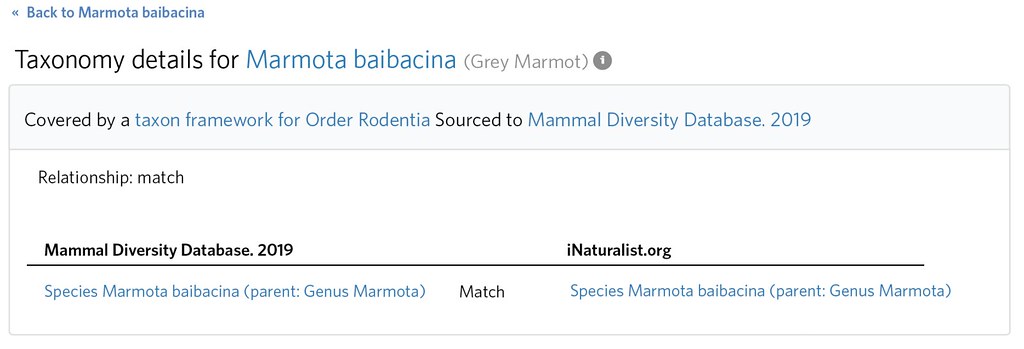
If the name, rank, parent name, and parent rank of the internal and external taxa are the same, iNaturalist will consider these a ‘Match’ (actually as long as the external taxon parent is an ancestor of the internal taxon it will count as a match) by creating a Taxon Framework Relationship connecting the taxa. Anything internal taxa covered by the taxon framework without a Taxon Framework Relationship are considered ‘Relationship unknowns’.

Unfortunately, to properly map these taxa, curators need to inspect these relationship unknown taxa and ideally either remove them (via a Taxon Change) or accomodate them with Deviations from the reference. Deviations are Taxon Framework Relationships other than Match. For example, if iNaturalist differs from the reference by splitting T. sierrae off from T. torosa, then a the Match Taxon Framework Relationship of T. torosa is misspecified since these taxa mean different things externally (sensu lato) and internally (sensu stricto). This Taxon Framework Relationship should be modified to include T. sierrae as a ‘one-to-many’. If T. novous is a ‘wholly new taxon’ (ie one that doesn’t influence what we mean by sister taxa) that is not in the reference but we wish to include in iNaturalist, then a ‘Not external’ Taxon Framework Relationship should be made. Deviations should include notes describing why the Deviation is necessary including any links to relevant flags.

A well curated Taxon Framework should have all taxa properly mapped to the External Reference. That is to say, there should be no internal taxa with unknown relationships.
There's a link to 'Relationship unknown' taxa from the taxonomy details page of any taxa defining a Taxon Framework. For example, there are many thousands for vascular plants. At the time of this writing, the rodent Marmota kastschenkoi had an unknown relationship.

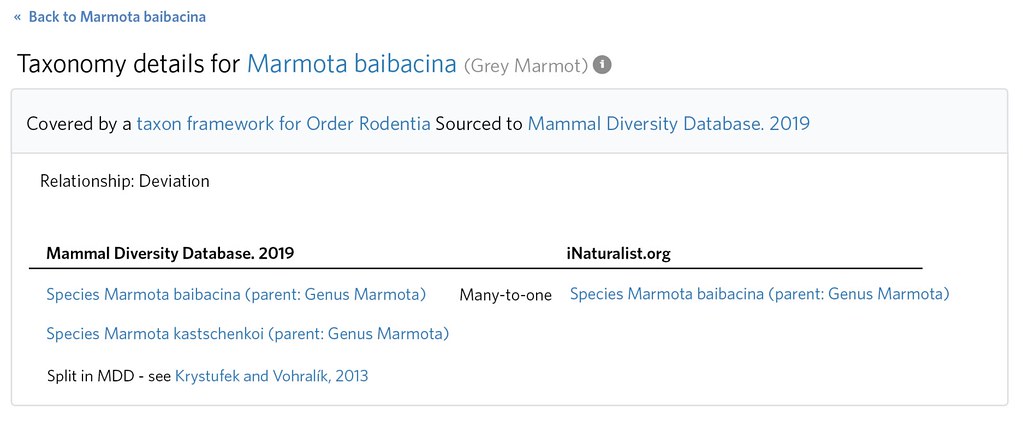
A little research reveals that Marmota kastschenkoi was previously considered a subspecies Marmota baibacina kastschenkoi and comprises the northern populations of Marmota baibacina 'sensu lato' (ie 'in the broad sense') .

Its clear that by Marmota baibacina, iNaturalist intends to means this broader 'sensu lato' concept that includes M. kastschenkoi. This is evident by inspecting the distribution (e.g. observations, taxon range map, atlas etc.) and also the taxonomy itself (e.g. Marmota baibacina kastschenkoi exists as a ssp of Marmota baibacina). This means that both the 'not internal' Taxon Framework Relationship for Marmota kastschenkoi and also the 'match' Taxon Framework Relationship for Marmota baibacina (since one is 'sensu lato' and one is in the narrow sense: 'sensu stricto' ) are misspecified.
The former should be deleted, and the latter should be expanded from a 'match' to a 'many to one' by editing the Taxon Framework Relationship and adding Marmota kastschenkoi as an additional 'external taxon'
There are cases when 'not internal' and 'not external' deviations are legitimate. They are used for obscure nodes (e.g. tribes) or ranks (e.g. hybrids) that might be missing from either iNaturalist or the external sources by design. But they also should be used for species 'wholly new to science'.
For example Svensson et al., 2013 describe a new galago from Angola. This species is sympatric with its closest relatives (G. thomasi and G. demidovii) and is much larger. This distinction between "a change in views of what a species is, that is, dependent on which species concept is adopted" and species "wholly new to science" is often blurry. When its not a clearcut case of the former, discerning when it is indeed the latter from the literature can be aggravating. This is made worse by the fact that journal articles tend to try to hype the 'new species discovery' parts of their paper for greater impact even if they are just reshuffling species. There's some nice text on the distinction in Svensson et al., 2013:
"Most new primate species recognized in the 21st century are the result of the taxonomic elevation of previously known subspecies to species (Groves, 2001; Macaca spp.: Kitchener & Groves, 2002; Aotus spp.: Defler & Bueno, 2007; Nomascus spp.: Thinh, Mootnick, Thanh, Nadler, & Roos, 2010; Microcebus spp.: Hotaling et al., 2016). Taxa wholly new to science, however, are also being described (e.g., Rungwecebus kipunji: Davenport et al., 2006; Jones et al., 2005; Tarsius tumpara: Shekelle, Groves, Merker, & Supriatna, 2008; Rhinopithecus strykeri: Geissmann et al., 2010; Nycticebus kayan: Munds et al., 2013)." Legitamite 'non-internal' Taxon Framework Relationships should include information on why the taxon is 'wholly new' and doesn't risk impacting neighbors (e.g. for Galagoides kumbirensis)
For iNaturalist's purposes. this distinction is important to the extent that we can:
1) avoid changes in the interpretation of past identifications
2) avoid associated meta-data with taxa (distribution information, default taxon photos, names etc.) from becoming misspecified
For example, in the Marmota kastschenkoi case, if we failed to alter the Taxon Relationship Framework mapping from 'non internal' to a 'many-to-one' relationship involving Marmota baibacina, then the proper coarse of action for adding Marmota kastschenkoi would be to create a new taxon. This would mean that the existing taxon range and atlas for Marmota baibacina would become misspecified (since they would continue being 'sensu lato' but the existence of Marmota kastschenkoi as a sibling would change the interpretation to 'sensu stricto'). Similarly, existing identifications of Marmota baibacina - which were intended to be interpreted as Marmota baibacina 'sensu lato' would start being interpreted as Marmota baibacina 'sensu stricto' and would result in the appearance of disagreements where none exist.
In contrast, by properly mapping Marmota kastschenkoi with a 'many-to-one' this would indicate that Marmota kastschenkoi should be added through a split and that additional work will be required to properly curate the distribution, identifications and explain to the community what the change means.
In the galago case, the new galago Galagoides kumbirensis can be simply added without any adverse impacts on its relatives.
Who can edit them?
Any taxon that is covered by a sourced Taxon Framework should have a Taxon Framework Relationship. Only curators can create, edit, or destroy Taxon Framework Relationships. If the Taxon Framework has Taxon Curators, only these taxon curators can create, edit or destroy them.
Good housekeeping of Taxon Framework Relationships
Generally, sourced Taxon Frameworks are initially populated through the back end of iNaturalist via a script that reads in the external taxa from the external reference and compares them to the internal iNaturalist taxa covered by the framework. The purpose of the script is to determine which species in iNaturalist aren't in the external reference (and vice versa) once all the deviations are taken into account. The script makes use of the Taxon Framework Relationships to do the mapping and will fail if there are issues with the Taxon Framework Relationships. Therefore its very important that curators take care when creating/editing/destroying Taxon Framework Relationships. Please do not leave loose ends around. For example, in curating Marmota kastschenkoi so that it is mapped through a many-to-one rather than a 'not internal' you need to do two steps (1) delete the 'not internal' Taxon Framework Relationship involving Marmota kastschenkoi and (2) edit the 'match' Taxon Framework Relationship involving Marmota baibacina by adding Marmota kastschenkoi to make it a 'many-to-one'. Just doing one of these two steps will corrupt the Taxon Framework Relationships.
Likewise, if you were to split Marmota baibacina (sensu lato) into Marmota baibacina (sensu stricto) and Marmota kastschenkoi, once the split is committed, remember your work is not done until you remove the many-to-one Taxon Framework Relationship on the inactive Marmota baibacina (sensu lato) and add new 'match' Taxon Framework Relationships to the outputs Marmota kastschenkoi and Marmota baibacina (sensu stricto).






